2010 - Transplantomics and Biomarkers in Transplantation
This page contains exclusive content for the member of the following sections: TTS. Log in to view.
MINI-ORAL PRESENTATIONS
4.7 - META-ANALYSIS OF SOLID ORGAN TRANSPLANT...
Presenter: Purvesh, Khatri, Stanford, USA
Authors: Purvesh Khatri, Richard Hayden Jones, Atul Butte, Minnie Sarwal.
META-ANALYSIS OF SOLID ORGAN TRANSPLANT DATA SETS IDENTIFIES DIFFERENTIALLY EXPRESSED MIRNAS COMMON IN HEART, KIDNEY AND LUNG ALLOGRAFTS
Purvesh Khatri, Richard Hayden Jones, Atul Butte, Minnie Sarwal. Department of Pediatrics, School of Medicine, Stanford University, Stanford, CA, USA.
Background: Recently, differentially expressed miRNAs in kidney and liver allograft rejection have been identified. The aim of our study was to investigate if there is a set of miRNAs that exhibit regulatory effects in acute rejection (AR), irrespective of the transplanted organ.
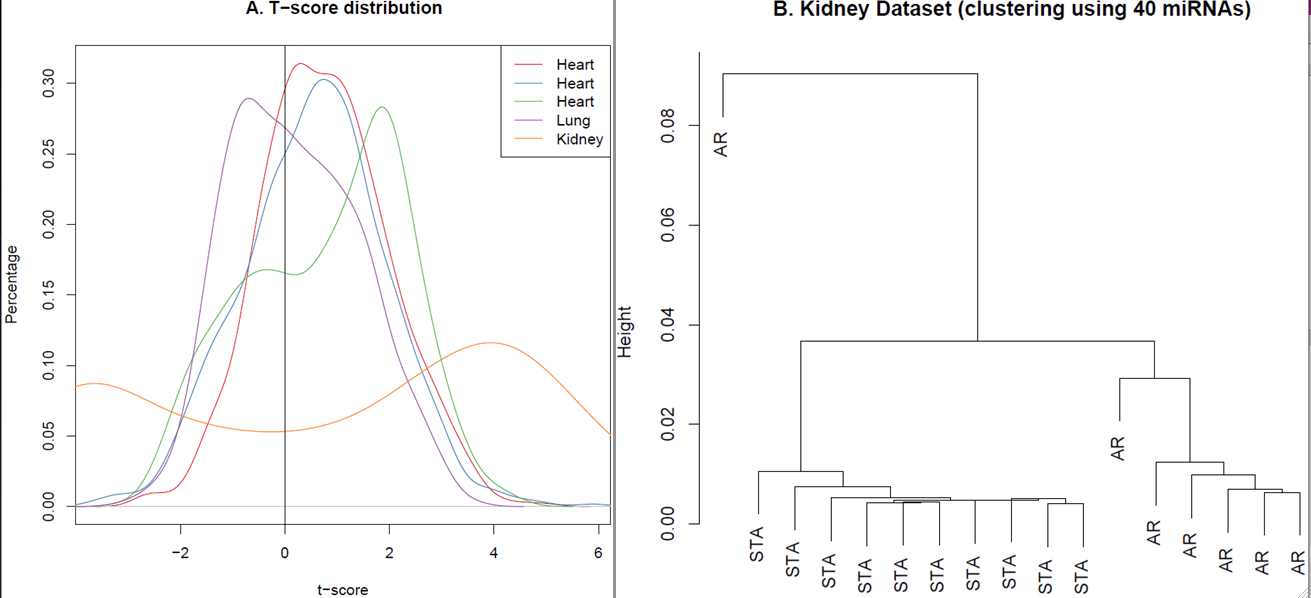
Method: We downloaded 5 allograft biopsy datasets from GEO (3 heart, 1 kidney and 1 lung datasets). Using miRBase, the genes in each sample in each dataset were divided into target and non-target genes. The genes in both groups were ranked by their expression values and difference in the average ranks for both groups was computed such that high difference in average ranks (called RE-score of a miRNA) indicate lower expression of target genes. We then calculated a t-score for each miRNA measuring the difference in RE-scores in AR versus stable (STA) samples, where higher t-score indicate higher regulatory effect of the corresponding miRNA in AR. The significance of t-scores was computed using sample label permutations for both two-tail and one-tail tests.
Results: All 5 datasets showed overall trend of higher t-scores (Fig. 1A), suggesting that many miRNAs have strong regulatory effects in AR. Using a two-tail test, we identified 40 miRNAs in the lung dataset, 278 miRNAs in the kidney and 40 miRNAs in all 3 heart datasets with statistically significant t-scores. Furthermore, there were 40 miRNAs that were statistically significant (FDR [lt] 0.2) in all 5 datasets. Interestingly, hierarchical clustering of the kidney dataset using the RE-scores of these 40 miRNAs classifies the samples into two distinct clusters with one sample misclassified (Fig. 1B). Similarly, using one-tail test, we identified 128 miRNAs with statistically significant positive t-scores in all 5 datasets. Interestingly, these 128 miRNAs include 9 miRNAs that were recently found to be differentially expressed in acute rejection of renal allografts, suggesting that they may also be differentially expressed during acute rejection in other solid organs .
Conclusion: Our analysis of solid organ transplant datasets suggests there may be a common set of miRNAs that play significant role in rejection of allografts.
Important Disclaimer
By viewing the material on this site you understand and accept that:
- The opinions and statements expressed on this site reflect the views of the author or authors and do not necessarily reflect those of The Transplantation Society and/or its Sections.
- The hosting of material on The Transplantation Society site does not signify endorsement of this material by The Transplantation Society and/or its Sections.
- The material is solely for educational purposes for qualified health care professionals.
- The Transplantation Society and/or its Sections are not liable for any decision made or action taken based on the information contained in the material on this site.
- The information cannot be used as a substitute for professional care.
- The information does not represent a standard of care.
- No physician-patient relationship is being established.
Contact
Address
The Transplantation Society
International Headquarters
740 Notre-Dame Ouest
Suite 1245
Montréal, QC, H3C 3X6
Canada
